Overview¶
pypillometry is a Python package for analyzing pupillometric data, providing a comprehensive toolkit for preprocessing, analyzing, and visualizing pupil size measurements and eye-tracking data. The package is built around the EyeData, PupilData, and GazeData classes, which serves as the main interface for handling pupillometric/eyetracking datasets.
PupilData: If you have only pupillometric data, use this class.GazeData: If you only have gaze data, use this class.EyeData: If you have both gaze and pupillometric data, use this class.
|
Class for handling eye-tracking data. |
|
Class representing Eye-Tracking data (x,y) from one or two eyes. |
|
Class representing pupillometric data. |
Generic class for eyedata. |
Importing eye-data¶
So far, the package provides functions to load data from Eyelink’s .edf files using the eyelinkio package via the function GenericEyeData.from_eyelink(). This function will import the raw data stored in the EDF file as well as all “message” triggers. It is also possible to use Python’s excellent functionality to parse text-based datafiles (most eyetrackers allow to export their proprietory format into a text-based one). For example, Eyelink’s ASC-format generated by the EDF2ASC conversion tool outputs space-separated data that can be easily loaded using the I/O functionality of the pandas package .
Once the data has been manually parsed into numpy-arrays, it can be converted into a PupilData, GazeData, or EyeData object.
|
Reads a |
|
Read a study from OSF using the configuration file. |
|
Read a study from a local directory using the configuration file. |
|
Authenticate against the OSF API using a personal access token. |
pypillometry provides functions to load data from local data directories (load_study_local()) and from OSF-projects (load_study_osf()) based on a user-provided configuration file (/examples/pypillometry_conf.py).
The package comes with a few example datasets for convenience (see this notebook). These can be loaded using the get_example_data() function.
Please refer to the following notebooks for more information:
Pipeline-based processing¶
pypillometry implements a pipeline-like approach where each operation executed on a PupilData,-GazeData or EyeData-object returns a reference to the (modified) object. This enables the “chaining” of commands as follows:
d=pp.get_example_data("rlmw_002_short").pupil_blinks_detect().blinks_merge_close().pupil_lowpass_filter(3).downsample(50)
This command loads an example data-file, applies a 3Hz low-pass filter to it, downsamples the signal to 50 Hz, detects blinks in the signal and merges short, successive blinks together. The final result of this processing-pipeline is stored in object d. This object stores also the complete history of the operations applied to the dataset and allows to transfer it to a new dataset.
By default, the operations are executed in place and the original object is modified. However, this can be changed by setting the inplace argument to False. This will return a new object with the results of the operations.
d=pp.get_example_data("rlmw_002_short")
d_new=d.pupil_blinks_detect(inplace=False).blinks_merge_close().pupil_lowpass_filter(3).downsample(50)
This will return a new object d_new with the results of the operations. The original object d is not modified.
See the following notebook for more on these issues:
Data modalities/variables and monocular vs. binocular data¶
pypillometry supports monocular and binocular data and allows to process several data modalities. The package ensures that all timeseries always have the same length/dimension such that all data modalities are aligned. The data are stored internally as an EyeDataDict-object which is a dictionary of numpy.ndarray-objects that are constrained to have the same shape. In addition, the keys of the dictionary are of the form “<eye>_<variable>”, e.g., left_pupil or right_x.
Most functions that operate on EyeData-objects allow to specify the eye and variable to operate on with optional arguments eye= and variable= (per default, all eyes and all variables are processed). Functions that can potentially operate on all modalities are implemented as methods of GenericEyeData while functions that are specific to pupillometric or eye-tracking data are implemented as methods of PupilData or GazeData. However, all objects be they PupilData, GazeData or EyeData inherit from GenericEyeData and therefore support the same set of operations.
Finally, new variables (or even “eyes”) can be created and added to the object. For example, when combining the data from both eyes, one can create a new variable that contains the average of the left and right pupil size which might be called “average_pupil”. In that case, the eye="average" and variable="pupil" arguments should be used to access this new variable.
Please see the following example for more details:
Plotting data¶
Each pypillometry object has a .plot attribute that gives access to the plotting functionality. Depending on the type of the object, the plotting functionality will be different. For example, GazeData objects (and also EyeData objects which inherit from that class) support the plotting of heatmaps or scanpaths.
Here is an overview of the plotting functionality:
|
Class for plotting pupil data. |
|
Class for plotting eye data. |
|
Analyzing pupillometric data¶
Analyzing eye-tracking data¶
work in progress
Artifact detection¶
The following is a list of functions designed for detecting artifacts. They all operate by detecting segments of suspicious data and either masking them in the data itself or returning them as Intervals objects for inspection.
|
Detect blinks in the pupillary signal using several strategies. |
|
Merge blinks that are close together. |
|
Calculate Euclidean distance between left and right eye coordinates and detect divergences. |
|
Detect and mask gaze coordinates that fall outside the screen boundaries. |
Each of these functions accepts a similar set of parameters:
eyes=/variables=: list of eyes/variables to process; if empty, all available eyes/variables are processed
apply_mask=: if True, the detected artifacts are applied as masks to the data and the object is returned; if False, the detected artifacts are returned as
Intervalsobjects.
A typical workflow would be to apply each of the functions to the data and tweak the parameters until the results are satisfactory. To do that, you might choose to set apply_mask=False and inspect the detected artifacts:
d=pp.get_example_data("rlmw_002_short")
blinks=d.pupil_blinks_detect(apply_mask=False)
print(blinks) # this is a dict with the blinks detected in each eye
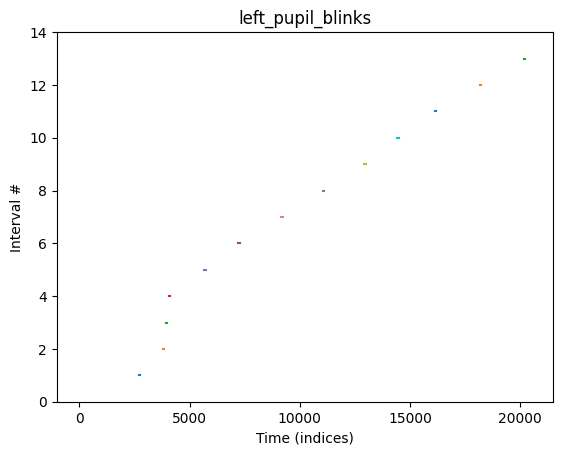
blinks["left_pupil"].plot() # plot when the blinks occurred over time (in the left eye)
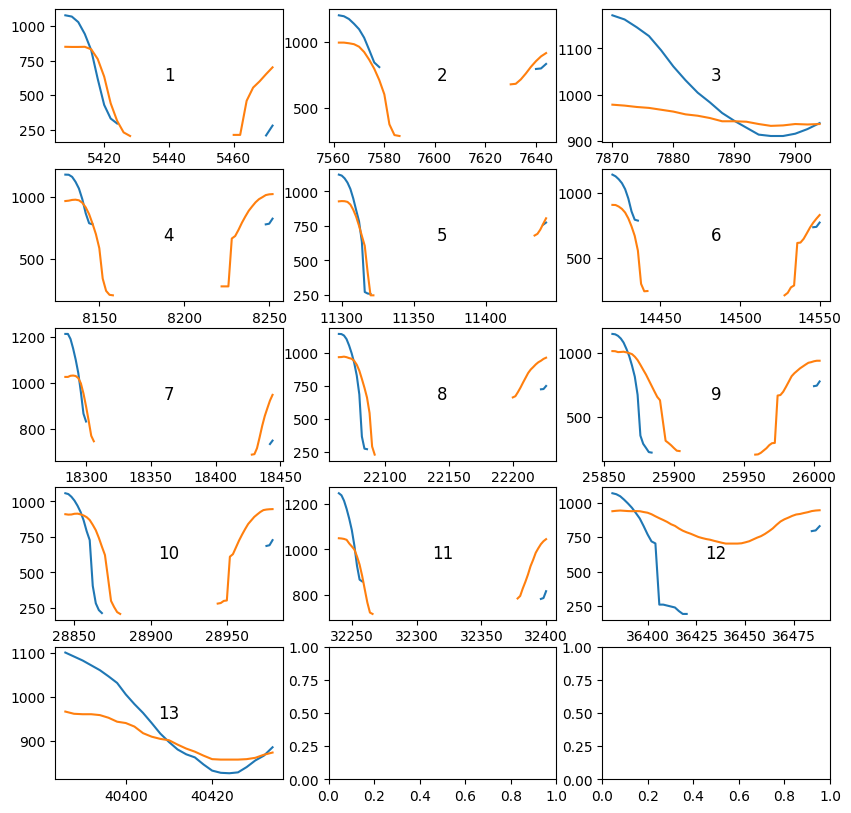
d.plot_intervals(blinks["left_pupil"], variables="pupil") # plot the data before and after the blinks
{'right_pupil': Intervals 'right_pupil_blinks' | 11 intervals (total duration: 662.00 None), 60.18 None +/- 16.04 None, [35.00 None, 81.00 None] | fs=500.0Hz, 'left_pupil': Intervals 'left_pupil_blinks' | 13 intervals (total duration: 892.00 None), 68.62 None +/- 24.26 None, [20.00 None, 98.00 None] | fs=500.0Hz}
[<Figure size 1000x1000 with 15 Axes>]
Finally, once you are satisfied, you can apply the masks to the data:
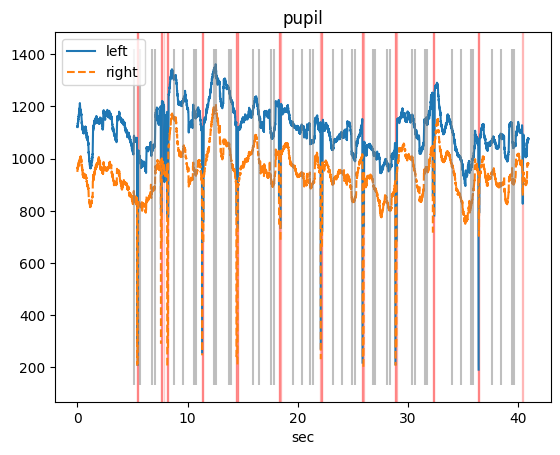
d.mask_intervals(blinks) # mask the data in the blinks
d.plot_timeseries(variables="pupil", units="sec") # plot the data after the blinks have been masked
Pre-processing data¶
Assuming you have generated a PupilData object, a range of pre-processing functions are available. The main pre-processing issues with pupillometric data are:
artifacts and missing data due to blinks (these can usually be corrected/interpolated)
missing data/artifacts from other sources (e.g., looking away, eyetracker losing pupil for other reasons)
smoothing/downsampling to get rid of high-freq low-amp noise
Handling Blinks¶
Pupillometric data usually contain blinks which show up as missing data in the signal where the eyetracker is unable to record the size of the pupil. A range of functions are available for detecting and interpolating blinks.
More details and an example can be found in the notebook: An example for how to handle blinks
A fully worked-out example of a real study can be found in this notebook: Preprocessing of a full dataset with multiple subjects
The following is a list of functions for that purpose. Note that the functions take multiple arguments that control the algorithms behaviour. It is often crucial to adjust the parameters on an individual level since the artifacts tend to be quite dissimilar between subjects (but usually stable within-subject). All arguments are documented in the API-docs.
|
Merge blinks that are close together. |
Smoothing/low-pass filtering¶
In most cases, pupillometric data should be low-pass filtered (e.g., using a cutoff of 4 Hz Jackson & Sirois, 2009) or smoothed in other ways (e.g., with a running-window).
Tge following is a list of functions for smoothing:
|
Simple downsampling scheme using mean within the downsampling window. |
Changing/Slicing data¶
Often, pupillometric data needs to be trimmed, e.g., to remove pre-experiment recordings or to remove unusable parts of the data (PupilData.sub_slice()). The timing should usually be realigned to the start of the experiment (PupilData.reset_time()). Furthermore, a scaling (e.g., Z-transform) of the pupil-data can be useful for comparing multiple subjects (PupilData.scale()).
The following is a list of available functions for these purposes:
|
Return a new EyeData object that is a shortened version of the current one (contains all data between start and end in units given by units (one of "ms", "sec", "min", "h"). |
|
Make and return a deep-copy of the pupil data. |
|
Scale the signal by subtracting mean and dividing by sd. |
|
Scale back to original values using either values provided as arguments or the values stored in scale_params. |
|
Resets time so that the time-array starts at time zero (t0). |
Plotting/Summarizing Data¶
Plotting¶
It is crucial to validate preprocessing steps by visually inspecting the results using plots. Therefore, pypillometry implements several plotting facilities that encourage active exploration of the dataset.
Please see the tutorial Plotting of pupillometric data for more details.
|
Inspecting/Summarizing¶
The package also provides several functions for summarizing datasets. Simply print()`ing a :class:`PupilData object gives a readable summary of the main properties of the dataset and also prints the complete history of the results. By calling PupilData.summary(), summary data can be arranged and summarized in tabular form.
See the notebook Summarizing pupillometric data for more details.
Return a summary of the dataset as a dictionary. |
|
|
Return result of applying a statistical function to data in a given interval relative to event-onsets. |
Modeling the pupillometric signal¶
For some applications, it is interesting to model the full pupillometric signal as consisting of a (tonic) baseline and a (phasic) response component. The package implements novel algorithms developed in our lab and documentation will become available here.
More details are availabel in this notebook: Modeling the pupillometric signal.
Artificial Data¶
For validation and testing purposes, it can be useful to generate artificial datasets. The package implements a FakePupilData as inheriting from regular PupilData and therefore shares all its functionality. In addition to that, FakePupilData stores “ground-truth” data and parameters that was used while creating the artificial data.
The function create_fake_pupildata() allows to quickly create datasets generating according to a provided experimental structure (see the functions documentation for an overview over the many available options).
